
| [ Information ] [ Publications ] [Signal processing codes] [ Signal & Image Links ] | |
| [ Main blog: A fortunate hive ] [ Blog: Information CLAde ] [ Personal links ] | |
| [ SIVA Conferences ] [ Other conference links ] [ Journal rankings ] | |
| [ Tutorial on 2D wavelets ] [ WITS: Where is the starlet? ] | |
If you cannot find anything more, look for something else (Bridget Fountain) |
|
|
|
|
Graph optimization methodologies, machine learning and artificial intelligence combined with biologically-related a-priori for genomic/transcriptomic/epigenetic data analysis and gene network inference


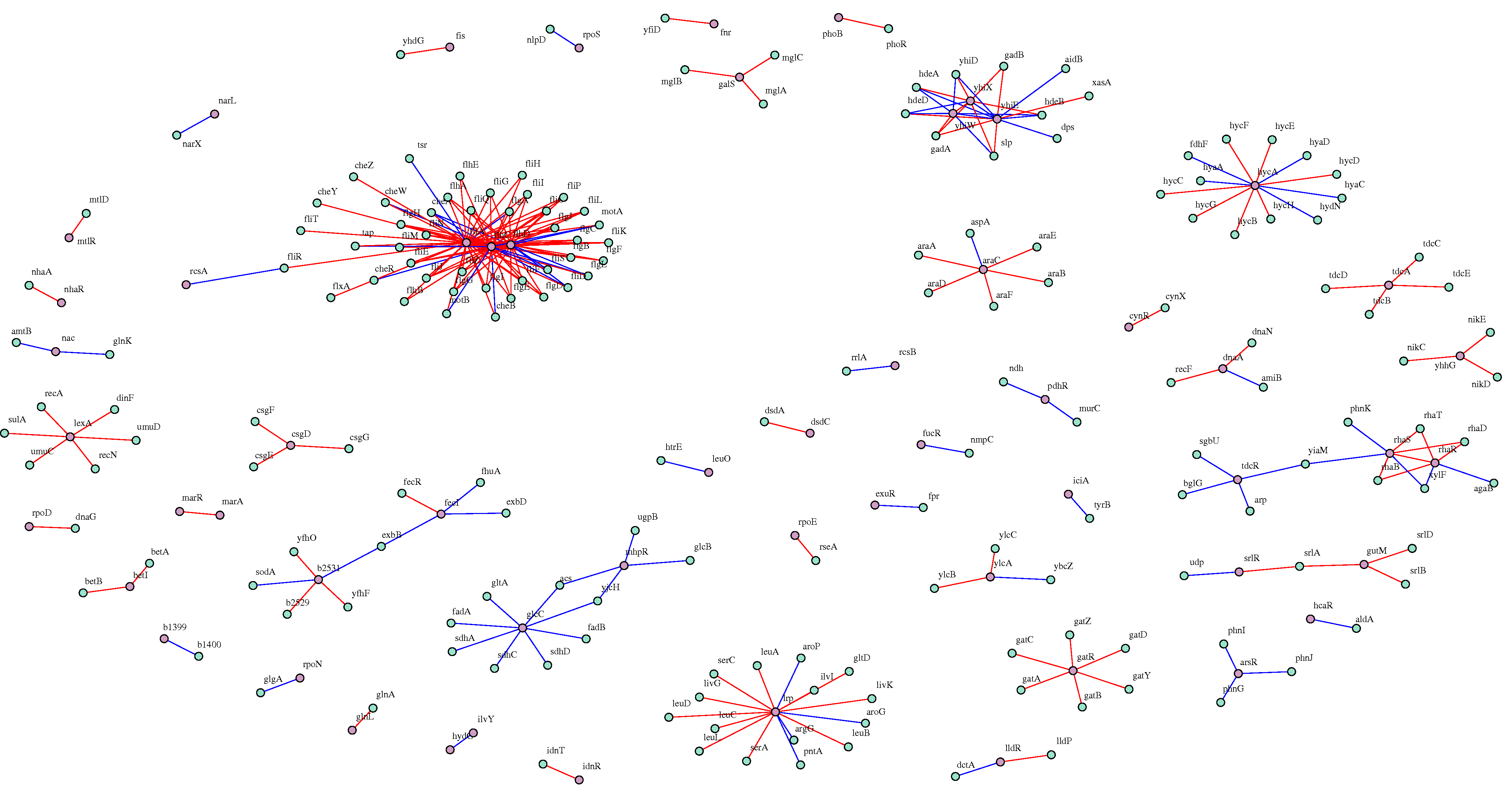
Discovering meaningful gene interactions is crucial for the identification of novel regulatory processes in cells. Building accurately the related graphs remains challenging due to the large number of possible solutions from available data. Nonetheless, enforcing a priori on the graph structure, such as modularity, may reduce network indeterminacy issues. BRANE Clust (Biologically-Related A priori Network Enhancement with Clustering) refines gene regulatory network (GRN) inference thanks to cluster information. It works as a post-processing tool for inference methods (i.e. CLR, GENIE3). In BRANE Clust, the clustering is based on the inversion of systems of linear equations involving a graph-Laplacian matrix promoting a modular structure. Our approach is validated on DREAM4 and DREAM5 datasets with objective measures, showing significant comparative improvements. We provide additional insights on the discovery of novel regulatory or co-expressed links in the inferred Escherichia coli network evaluated using the STRING database. The comparative pertinence of clustering is discussed computationally (SIMoNe, WGCNA, X-means) and biologically (RegulonDB). BRANE Clust software is available at: http://www-syscom.univ-mlv.fr/~pirayre/Codes-GRN-BRANE-clust.html.
Keywords: Optimization, Labeling, Biological information theory, Graphical models, Probabilistic logic, Merging, Databases, Gene regulatory network, clustering, combinatorial optimization, transcriptomic data, DREAM challenge

Background Inferring gene networks from high-throughput data (RNA-Seq) constitutes an important step in the discovery of relevant regulatory relationships in organism cells. Despite the large number of available Gene Regulatory Network inference methods, the problem remains challenging: the underdetermination in the space of possible solutions requires additional constraints that incorporate a priori information on gene interactions.
Results Weighting all possible pairwise gene relationships by a probability of edge presence, we formulate the regulatory network inference as a discrete variational problem on graphs. We enforce biologically plausible coupling between groups and types of genes by minimizing an edge labeling functional coding for a priori structures. The optimization is carried out with Graph cuts, an approach popular in image processing and computer vision. We compare the inferred regulatory networks to results achieved by the mutual-information-based Context Likelihood of Relatedness (CLR) method and to the state-of-the-art GENIE3, winner of the DREAM4 multifactorial challenge.
Conclusions Our BRANE Cut approach infers more accurately the five DREAM4 in silico networks (with improvements from 6% to 11%). On a real Escherichia coli compendium, an improvement of 11.8% compared to CLR and 3% compared to GENIE3 is obtained in terms of Area Under Precision-Recall curve. Up to 48 additional verified interactions are obtained over GENIE3 for a given precision. On this dataset involving 4345 genes, our method achieves a performance similar to that of GENIE3, while being more than seven times faster. The BRANE Cut code is available at: http://www-syscom.univ-mlv.fr/~pirayre/Codes-GRN-BRANE-cut.html
Keywords: Gene network inference, high throughput data, optimization, network theory, maximum flow