| [ Information ] [ Publications ] [Signal processing codes] [ Signal & Image Links ] | |
| [ Main blog: A fortunate hive ] [ Blog: Information CLAde ] [ Personal links ] | |
| [ SIVA Conferences ] [ Other conference links ] [ Journal rankings ] | |
| [ Tutorial on 2D wavelets ] [ WITS: Where is the starlet? ] | |
If you cannot find anything more, look for something else (Bridget Fountain) |
|
|
|
|
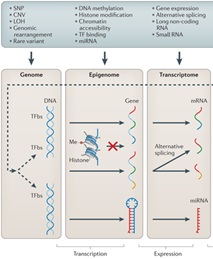
In its commitment to successfully carry out the energy transition, IFP Energies nouvelles is conducting research to optimize biotechnological processes with applications to more renewable energy sources. These processes require the use of microorganisms, for which we need to deepen our understanding of their molecular mechanisms. In our approach, we adopt a systemic approach that considers different levels of biological regulation that interact with each other. We have gathered a set of genetic data, information on gene activity, and epigenetic imprints for our model organism Trichoderma reesei. But the question that arises is: how can we detect differences in the functioning of a biological system by combining different experimental data? To answer this question, we want to develop and implement new methodologies that integrate different types of data by identifying both the fundamental systemic mechanisms that remain constant and those specific to each experimental condition. We propose to explore different statistical analysis, data processing and optimization approaches such as Bayesian methods, source separation, and deep learning through variational autoencoders. These tools will help us better understand the functioning of our microorganisms of interest to optimize biotechnological processes in the fields of bio-based chemistry and biofuels. This thesis is linked to the targeted project GalaxyBioProd of the PEPR B-BEST --- Biomasse, biotechnologies, technologies pour la chimie verte et les énergies renouvelables --- (PEPR, Programmes et Équipements Prioritaires de Recherche or Priority Research Programmes and Equipments, are aimed at constructing or consolidating French leadership in specific scientific fields). The tools developed in this thesis will be made available to the community of biologists through their integration into the Galaxy platform.
We are looking for a motivated student with strong skills in statistics, machine learning and bioinformatics. Prior experience in processing and analyzing omics data and computer programming is highly desirable. The candidate will work closely with our team of researchers and will benefit from a stimulating environment conducive to learning and professional development. Send resume and motivated application letter to:
